
 |
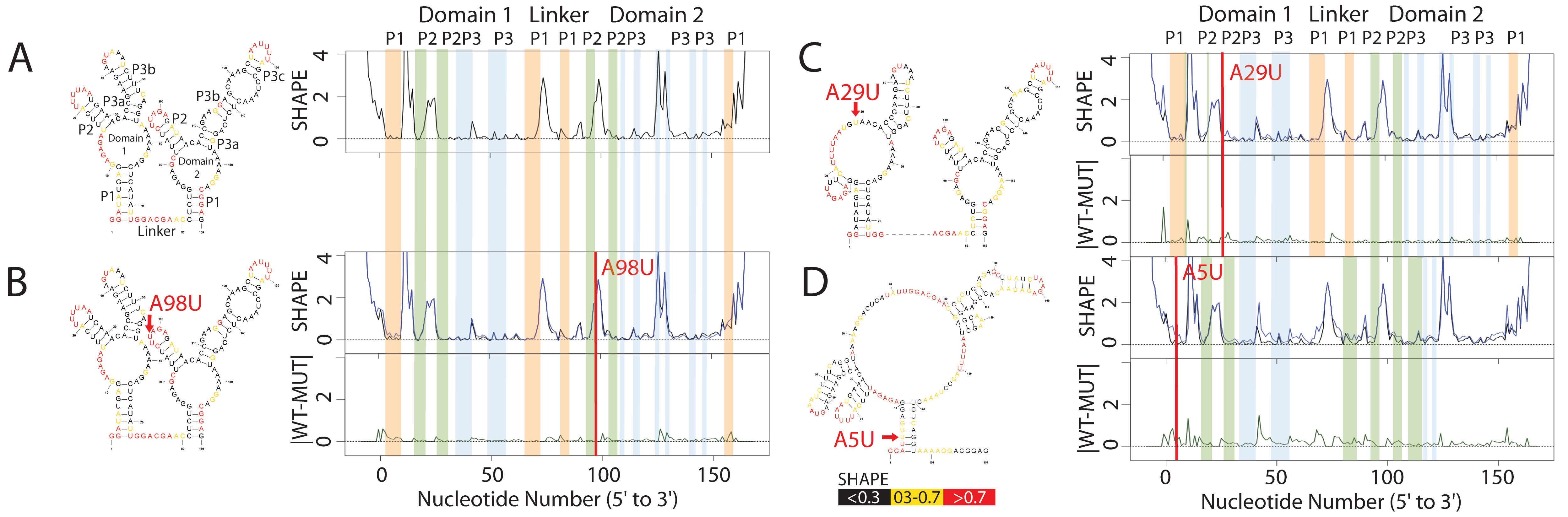 AboutMutations (or Single Nucleotide Variants) in folded RiboNucleic Acid structures that cause local or global conformational change are riboSNitches. Predicting riboSNitches is challenging as it requires making two, albeit related, structure predictions. The data most often used to experimentally validate riboSNitch predictions is Selective 2’ Hydroxyl Acylation by Primer Extension, or SHAPE. Experimentally establishing a riboSNitch requires the quantitative comparison of two SHAPE traces: wild-type (WT) and mutant. Historically, SHAPE data was collected on electropherograms and change in structure was evaluated by “gel gazing.” SHAPE data is now routinely collected with next generation sequencing and/or capillary sequencers. We have established a classifier capable of simulating human “gazing” by identifying features of the SHAPE profile that human experts agree “looks” like a riboSNitch. |
Authors |
Chanin Tolson and Alain Laederach |
Download |
Package source (use this for Linux / Solaris): classSNitch_1.0.0.tar.gz Windows binary: classSNitch_1.0.0.zip Repository: classSNitch is also available on R-Forge |
Installation |
Linux: At the command line type "R CMD INSTALL classSNitch_1.0.0.tar.gz". Windows: From the pull-down menu, click on "Packages->Install Packages from local zip file", and then select the downloaded file docs/classSNitch_1.0.0.zip. R install: install.packages("classSNitch", repos="http://R-Forge.R-project.org", type="source") Make sure R is installed on your computer. It is freely available on R-Forge You will also need randomForest, dtw, ROCR, and gplots packages (available from the Comprehensive R Archive Network). |
Getting Started |
Load classSNitch package: library("classSNitch") Get list of help pages: help(package="classSNitch") Documentation: Reference Manual |
Examples |
#get change parameters data("shape_ex") sample_shape = shape_ex sample = getChangeParams(sample_shape[2:nrow(sample_shape),], base=sample_shape[1,], trim=5) #predict change cr = classifyRNA(classes=2) cr_pred = predict(cr, sample, resp="response") |
Contact |
Send an email to the Laederach Lab. We welcome any questions, comments or suggestions. If you have any publications using classSNitch, we would be interested in seeing how the package is being used. |